15.2.7 Algorithm (Multiple Linear Regression)Multi-Regression-Algorithm
The Multiple Linear Regression Model
Multiple Linear Regression Model
Multiple linear regression is an extension of the simple linear regression where multiple independent variables exist. It is used to analyze the effect of more than one independent variable 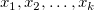 on the dependent variable y. For a given dataset on the dependent variable y. For a given dataset 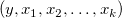 , the multiple linear regression fits the dataset to the model: , the multiple linear regression fits the dataset to the model:
where 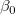 is the y-intercept and the parameters is the y-intercept and the parameters 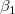 , , 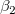 , ... , , ... , 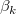 are called the partial coefficients.
It can be written in matrix form: are called the partial coefficients.
It can be written in matrix form:
where
Assuming that 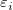 are independent and identically distributed as normal random variables with are independent and identically distributed as normal random variables with 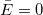 and and ![Var[E]=\sigma^2 Var[E]=\sigma^2](//d2mvzyuse3lwjc.cloudfront.net/doc/en/UserGuide/images/Multiple_Regression_Results/math-5d3cf3883ed3c4d0b917f9fc2e8f776b.png?v=0) .
In Order to minimize the .
In Order to minimize the 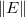 with respect to with respect to 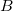 , we solve the function: , we solve the function:
The results 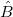 is the least square estimate of the vector B, and it is the solution to the linear equations, which can be expressed as: is the least square estimate of the vector B, and it is the solution to the linear equations, which can be expressed as:
where X' is the transpose of X.
The predicted value of Y for a given X is:
By substituting 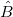 into (4), we can and defined matrix into (4), we can and defined matrix 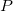 . .
The residuals is defined as:
and the residual sum of squares can be written by:
Fit Control
Errors as Weight
We can give weight to each 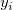 in fitting process, the yEr± error column in fitting process, the yEr± error column 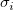 is treated as weight is treated as weight 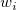 for each for each 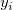 , when yEr± is abscent, , when yEr± is abscent, 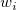 should be 1 for all should be 1 for all  . .
The solution 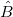 for fitting with weight can be written as: for fitting with weight can be written as:
where
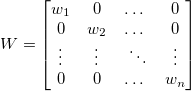
No Weighting
The error bar will not be treated as weight in calculation.
Direct Weighting
Instrumental
Fix Intercept (at)
Fix intercept will set the y-intercept 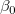 to a fixed value, meanwhile, the total degree of freedom will be n*=n-1 due to the intercept fixed. to a fixed value, meanwhile, the total degree of freedom will be n*=n-1 due to the intercept fixed.
Scale Error with sqrt(Reduced Chi-Sqr)
Scale Error with sqrt(Reduced Chi-Sqr) is available when fitting with weight. This option only affects the error on the parameters reported from the fitting process, and does not affect the fitting process or the data in any way.
By default, it is checked, and 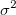 , which is the variance of , which is the variance of 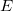 is taken into account for calculating error on the parameters, otherwise, variance of is taken into account for calculating error on the parameters, otherwise, variance of 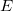 will not be taken into account for error calculation.
Take Covariance Matrix as example: will not be taken into account for error calculation.
Take Covariance Matrix as example:
Scale Error with sqrt(Reduced Chi-Sqr)
Do not Scale Error with sqrt(Reduced Chi-Sqr)
For weighted fitting, 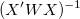 is used instead of is used instead of 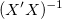 . .
Fitting Results
Fit Parameters

The Fitted Values
Formula (4)
The Parameter Standard Errors
For each parameter, the standard error can be obtained by:
where 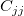 is the jth diagonal element of is the jth diagonal element of 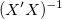 (note that (note that 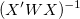 is used for weight fitting). The residual standard deviation is used for weight fitting). The residual standard deviation 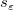 (also called "std dev", "standard error of estimate", or "root MSE") is computed as: (also called "std dev", "standard error of estimate", or "root MSE") is computed as:
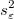 is an estimate of is an estimate of 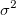 , which is variance of , which is variance of 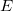
| Note: Please read the ANOVA Table for more details about the degree of freedom (df), dfError.
|
t-Value and Confidence Level
If the regression assumptions hold, we can perform the t-tests for the regression coefficients with the null hypotheses and the alternative hypotheses:
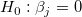
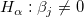
The t-values can be computed as:
With the t-value, we can decide whether or not to reject the corresponding null hypothesis. Usually, for a given Confidence Level for Parameters:  , we can reject , we can reject 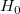 when when 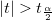 . Additionally, the p-value is less than . Additionally, the p-value is less than  . .
Prob>|t|
The probability that 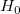 in the t test is true. in the t test is true.
where 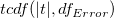 computes the cumulative distribution function of the Student's t distribution at the values |t|, with degree of freedom of error computes the cumulative distribution function of the Student's t distribution at the values |t|, with degree of freedom of error 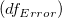 . .
LCL and UCL
From the t-value, we can calculate the 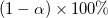 Confidence Interval for each parameter by: Confidence Interval for each parameter by:
where 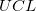 and and 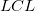 is short for the Upper Confidence Interval and Lower Confidence Interval, respectively. is short for the Upper Confidence Interval and Lower Confidence Interval, respectively.
CI Half Width
The Confidence Interval Half Width is:
VIF
The Variance Inflation Factor is:
Where: 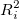 is unadjusted coefficient of determination for regression the ith independent variable on the remaining ones. is unadjusted coefficient of determination for regression the ith independent variable on the remaining ones.
Fit Statistics
Some fit statistics formulas are summary here:
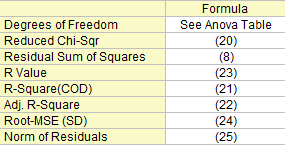
Degree of Freedom
The degree of freedom for (Error) variation. Please refer to the ANOVA table for more details.
Reduced Chi-Sqr
Residual Sum of Squares
The residual sum of squares, see formula (8).
R-Square (COD)
The goodness of fit can be evaluated by Coefficient of Determination (COD), 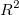 , which is given by: , which is given by:
Adj. R-Square
The adjusted 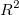 is used to adjust the is used to adjust the 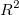 value for the degree of freedom. It can be computed as: value for the degree of freedom. It can be computed as:
R Value
Then we can compute the R-value, which is simply the square root of 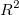 : :
Root-MSE (SD)
Root Mean Square of the Error, or residual standard deviation, which equals to:
Norm of Residuals
Equals to square root of RSS:
ANOVA Table
The ANOVA table of linear fitting is:
|
|
df
|
Sum of Squares
|
Mean Square
|
F Value
|
Prob > F
|
| Model
|
k
|
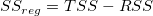
|
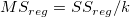
|
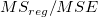
|
p-value
|
| Error
|
n* - k
|
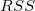
|
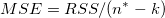
|
|
|
| Total
|
n*
|
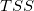
|
|
|
|
| Note: If intercept is included in the model, n*=n-1. Otherwise, n*=n and the total sum of squares is uncorrected.
|
Where the total sum of square, TSS, is:
The F value here is a test of whether the fitting model differs significantly from the model y=constant.
Additionally, the p-value, or significance level, is reported with an F-test. We can reject the null hypothesis if the p-value is less than  , which means that fitting model differs significantly from the model y=constant. , which means that fitting model differs significantly from the model y=constant.
If fixing the intercept at a certain value, the p value for F-test is not meaningful, and it is different from that in multiple linear regression without the intercept constraint.
Lack of fit table
To run the lack of fit test, you need to have repeated observations, namely, "replicate data" , so that at least one of the X values is repeated within the dataset, or within multiple datasets when concatenate fit mode is selected.
Notations used for fit with replicates data:
The sum of square in table below is expressed by:
The Lack of fit table of linear fitting is:
|
|
DF
|
Sum of Squares
|
Mean Square
|
F Value
|
Prob > F
|
| Lack of Fit
|
c-k-1
|
LFSS
|
MSLF = LFSS / (c - k - 1)
|
MSLF / MSPE
|
p-value
|
| Pure Error
|
n - c
|
PESS
|
MSPE = PESS / (n - c)
|
|
|
| Error
|
n*-k
|
RSS
|
|
|
|
Note:
If intercept is included in the model, n*=n-1. Otherwise, n*=n and the total sum of squares is uncorrected. If the slope is fixed, 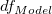 = 0. = 0.
c denotes the number of distinct x values. If intercept is fixed, DF for Lack of Fit is c-k.
|
Covariance and Correlation Matrix
The covariance matrix for the multiple linear regression can be calculated as
The correlation between any two parameters is:
Residual Analysis
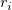 stands for the Regular Residual stands for the Regular Residual 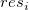 . .
Standardized
Studentized
Also known as internally studentized residual.
Studentized deleted
Also known as externally studentized residual.
In the equations for the Studentized and Studentized deleted residuals, 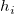 is the ith diagonal element of the matrix is the ith diagonal element of the matrix 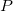 : :
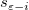 means the variance is calculated based on all points but exclude the ith. means the variance is calculated based on all points but exclude the ith.
Plots
Partial Leverage Plots
In multiple regression, partial leverage plots can be used to study the relationship between the independent variable and a given dependent variable. In the plot, the partial residual of Y is plotted against the partial residual of X, or the intercept. The partial residual of a certain variable is the regression residual with that variable omitted in the model.
Take the model 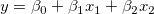 for example: the partial leverage plot for for example: the partial leverage plot for 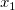 is created by plotting the regression residual of is created by plotting the regression residual of 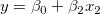 against the residual of against the residual of 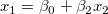 . .
Resudial Type
Select one residual type among Regular, Standardized, Studentized, Studentized Deleted for Plots.
Residual vs. Independent
Scatter plot of residual 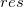 vs. indenpendent variable vs. indenpendent variable 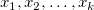 , each plot is locate in a seperate graphs. , each plot is locate in a seperate graphs.
Residual vs. Predicted Value
Scatter plot of residual 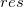 vs. fitted results vs. fitted results 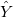 . .
Residual vs. Order of the Data
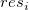 vs. sequence number vs. sequence number 
Histogram of the Residual
The Histogram plot of the Residual 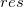
Residual Lag Plot
Residuals 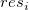 vs. lagged residual vs. lagged residual 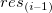 . .
Normal Probability Plot of Residuals
A normal probability plot of the residuals can be used to check whether the variance is normally distributed as well. If the resulting plot is approximately linear, we proceed to assume that the error terms are normally distributed. The plot is based on the percentiles versus ordered residual, the percentiles is estimated by
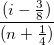
where n is the total number of dataset and i is the i th data. Also refer to Probability Plot and Q-Q Plot
|